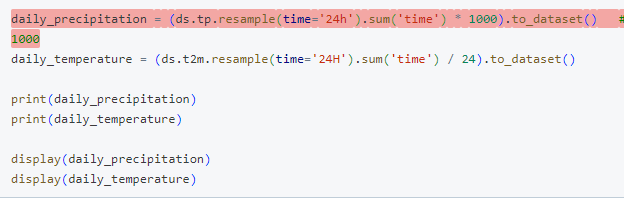
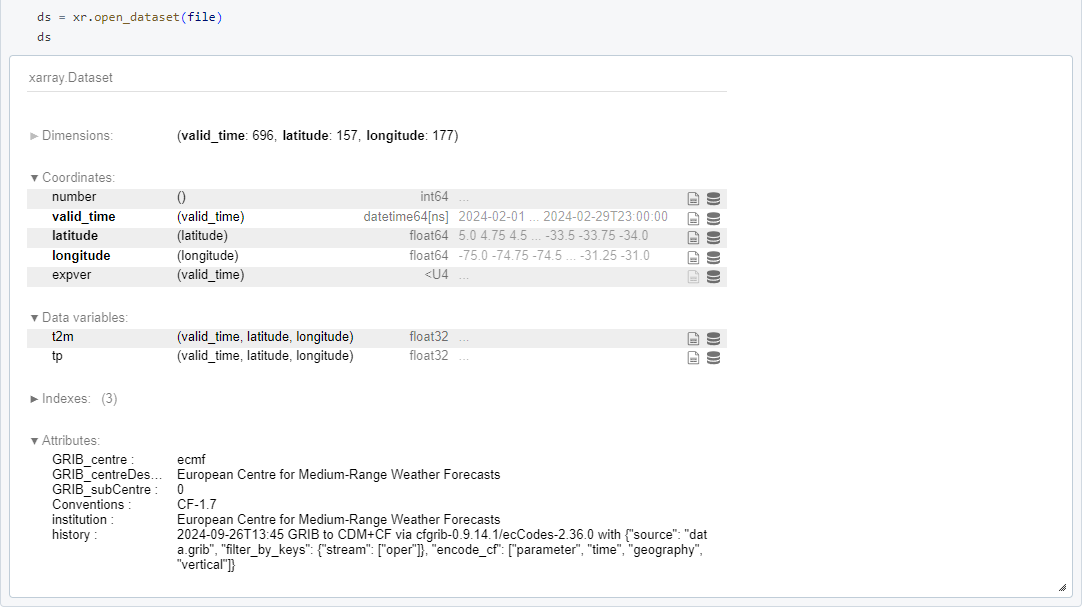
Thanks Milana, I didn't think about the spatial distribution, that must be it!
Vivien MAVEL Milana Vuckovicthanks, I really appreciate your answers.
As Milana Vuckoviccommented, ERA5 land data contains information just for land regions.
The data I use was downloaded considering a goegraphic coordinates covering some ocean zones. I employ a western South America area, particularly which geographic coordinates are -83 (west), -23 (south), -67 (east), and 3 (north).
Tentative explanation of RuntimeWarning: Mean of empty slice return np.nanmean(a, axis=axis, dtype=dtype) can be the missing values for ocean zones. In order to be sure of it, it is neccesary to test it.
It is suggested two options: 1) try with a land area exclusively or 2) graph the data.
Regarding second option, I explored in different forums trying to know how to plot an specific area (using netCDF file downloaded from ERA 5 land) but so far I did not find a way to do it. If you know how to do it, it will help me a lot!
Hi Brian,
This is how you select smaller area using xarray:
small_area = era5_land.sel(latitude=slice(-10,-20),longitude=slice(10,20)) Note that latitudes are going from bigger to smaller because they decrease in your dataset too.
If you need the data over the oceans maybe you should better use ERA5 instead of ERA5 Land.
Before explaining the RuntimeWarning I have tested it both on ERA5 and ERA5 Land and on smaller area, and I got the warning with ERA5 Land which then disappeared when I selected smaller area. Your daily mean will be calculated fine, it is just telling you that you don't have the data over the ocean which you already know.
Milana Vuckovic I just tried what you said in small area (covering only land region), and it works. The "RuntimeWarning" does not appear anymore. Thanks
For a CDO-based solution, the relevant command is shifttime, which essentially does what is says on the can and shifts the time stamp.
This kind of problem arises frequently with any kind of flux or accumulated field where the timestep points to the END of the time "window", for example, with 3 hourly TRMM data the last three hours of the day have the stamp of 00 on the date afterwards.
So for ERA5 precip where you have a long series of hourly data from ERA5, and the window from 23:00 to 00:00 has the timestamp of 00:00 from the day after, and you want the daily average, you can do:
cdo shifttime,-1hour in.nc shift.nc # now step 0 on Jan 2 has Jan 1, 23:00 stamp
cdo daymean shift.nc daymean.nc or piped together:
cdo daymean -shifttime,-1hour in.nc daymean.ncHi, Adrian Mark Tompkins I have applied your code and got the following result.
cdo sinfo d2m_wb.nc
File format : NetCDF2
-1 : Institut Source T Steptype Levels Num Points Num Dtype : Parameter ID
1 : unknown unknown v instant 1 1 475 1 F64 : -1
Grid coordinates :
1 : lonlat : points=475 (19x25)
lon : 85.5 to 90 by 0.25 degrees_east
lat : 21.5 to 27.5 by 0.25 degrees_north
Vertical coordinates :
1 : surface : levels=1
Time coordinate : 25904 steps
RefTime = 1900-01-01 00:00:00 Units = hours Calendar = gregorian Bounds = true
YYYY-MM-DD hh:mm:ss YYYY-MM-DD hh:mm:ss YYYY-MM-DD hh:mm:ss YYYY-MM-DD hh:mm:ss
1949-12-31 23:00:00 1950-01-01 11:00:00 1950-01-02 11:00:00 1950-01-03 11:00:00
1950-01-04 11:00:00 1950-01-05 11:00:00 1950-01-06 11:00:00 1950-01-07 11:00:00
1950-01-08 11:00:00 1950-01-09 11:00:00 1950-01-10 11:00:00 1950-01-11 11:00:00
1950-01-12 11:00:00 1950-01-13 11:00:00 1950-01-14 11:00:00 1950-01-15 11:00:00
1950-01-16 11:00:00 1950-01-17 11:00:00 1950-01-18 11:00:00 1950-01-19 11:00:00
1950-01-20 11:00:00 1950-01-21 11:00:00 1950-01-22 11:00:00 1950-01-23 11:00:00
1950-01-24 11:00:00 1950-01-25 11:00:00 1950-01-26 11:00:00 1950-01-27 11:00:00
1950-01-28 11:00:00 1950-01-29 11:00:00 1950-01-30 11:00:00 1950-01-31 11:00:00
1950-02-01 11:00:00 1950-02-02 11:00:00 1950-02-03 11:00:00 1950-02-04 11:00:00
1950-02-05 11:00:00 1950-02-06 11:00:00 1950-02-07 11:00:00 1950-02-08 11:00:00
1950-02-09 11:00:00 1950-02-10 11:00:00 1950-02-11 11:00:00 1950-02-12 11:00:00
1950-02-13 11:00:00 1950-02-14 11:00:00 1950-02-15 11:00:00 1950-02-16 11:00:00
1950-02-17 11:00:00 1950-02-18 11:00:00 1950-02-19 11:00:00 1950-02-20 11:00:00
1950-02-21 11:00:00 1950-02-22 11:00:00 1950-02-23 11:00:00 1950-02-24 11:00:00
1950-02-25 11:00:00 1950-02-26 11:00:00 1950-02-27 11:00:00 1950-02-28 11:00:00
................................................................................
................................................................................
................................................................................
.................
2020-10-03 11:00:00 2020-10-04 11:00:00 2020-10-05 11:00:00 2020-10-06 11:00:00
2020-10-07 11:00:00 2020-10-08 11:00:00 2020-10-09 11:00:00 2020-10-10 11:00:00
2020-10-11 11:00:00 2020-10-12 11:00:00 2020-10-13 11:00:00 2020-10-14 11:00:00
2020-10-15 11:00:00 2020-10-16 11:00:00 2020-10-17 11:00:00 2020-10-18 11:00:00
2020-10-19 11:00:00 2020-10-20 11:00:00 2020-10-21 11:00:00 2020-10-22 11:00:00
2020-10-23 11:00:00 2020-10-24 11:00:00 2020-10-25 11:00:00 2020-10-26 11:00:00
2020-10-27 11:00:00 2020-10-28 11:00:00 2020-10-29 11:00:00 2020-10-30 11:00:00
2020-10-31 11:00:00 2020-11-01 11:00:00 2020-11-02 11:00:00 2020-11-03 11:00:00
2020-11-04 11:00:00 2020-11-05 11:00:00 2020-11-06 11:00:00 2020-11-07 11:00:00
2020-11-08 11:00:00 2020-11-09 11:00:00 2020-11-10 11:00:00 2020-11-11 11:00:00
2020-11-12 11:00:00 2020-11-13 11:00:00 2020-11-14 11:00:00 2020-11-15 11:00:00
2020-11-16 11:00:00 2020-11-17 11:00:00 2020-11-18 11:00:00 2020-11-19 11:00:00
2020-11-20 11:00:00 2020-11-21 11:00:00 2020-11-22 11:00:00 2020-11-23 11:00:00
2020-11-24 11:00:00 2020-11-25 11:00:00 2020-11-26 11:00:00 2020-11-27 11:00:00
2020-11-28 11:00:00 2020-11-29 11:00:00 2020-11-30 11:00:00 2020-12-31 23:00:00
cdo sinfo: Processed 1 variable over 25904 timesteps [6.03s 37MB
In this case, the timestep shows 11:00:00. I guess it should be 12:00:00. What wrong I've done here? Any suggestion? Thank you.
I know it is old post but any idea how this will work with beta api I am getting KeyError: time
cds = cdsapi.Client()
dataset = “reanalysis-era5-single-levels”
params = {
“data_format”: “netcdf”,
‘product_type’: ‘reanalysis’,
‘variable’: [‘2m_temperature’,‘total_precipitation’],
‘year’: year,
‘month’: month,
‘day’: days,
‘time’:
[
‘00:00’, ‘01:00’, ‘02:00’,
‘03:00’, ‘04:00’, ‘05:00’,
‘06:00’, ‘07:00’, ‘08:00’,
‘09:00’, ‘10:00’, ‘11:00’,
‘12:00’, ‘13:00’, ‘14:00’,
‘15:00’, ‘16:00’, ‘17:00’,
‘18:00’, ‘19:00’, ‘20:00’,
‘21:00’, ‘22:00’, ‘23:00’,
],
‘area’: [5, -75, -34, -31]
}
file_path = cds.retrieve(dataset, params)