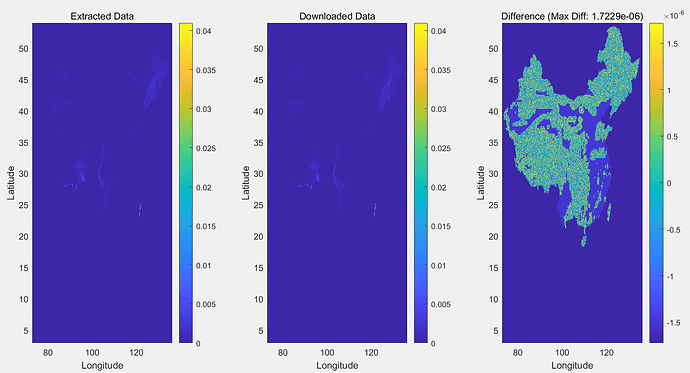
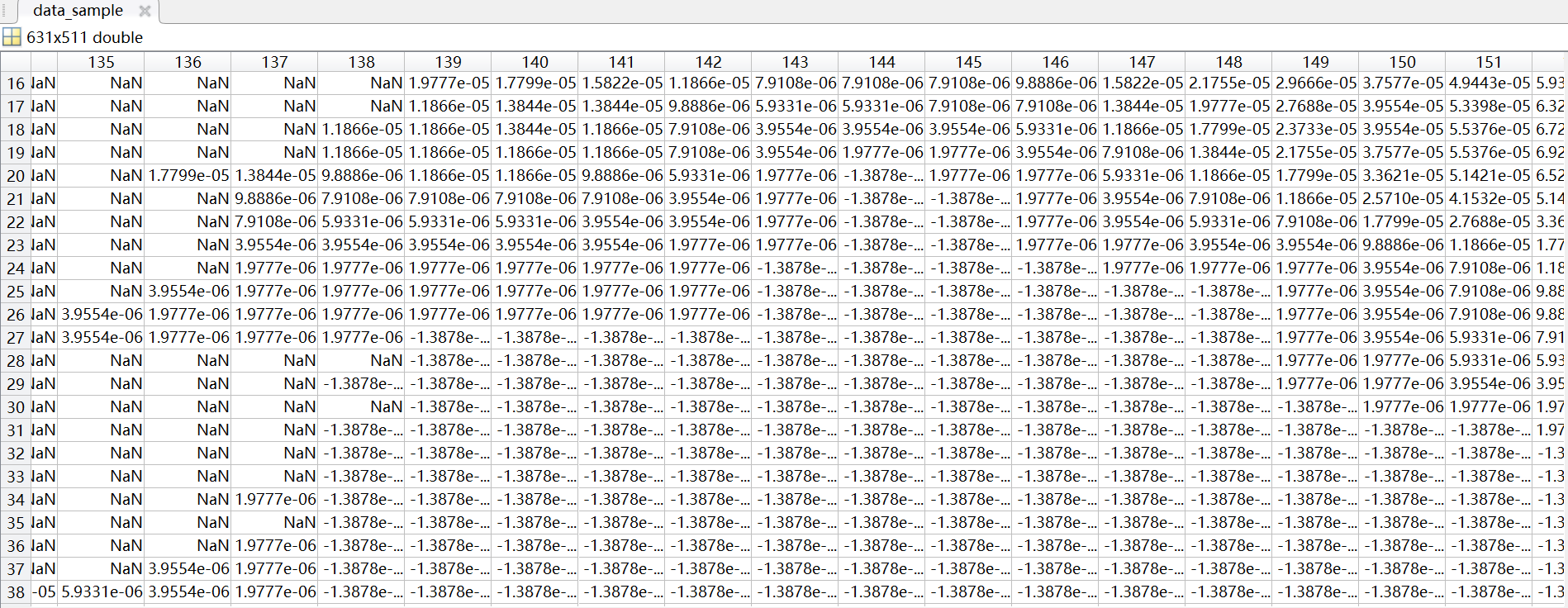
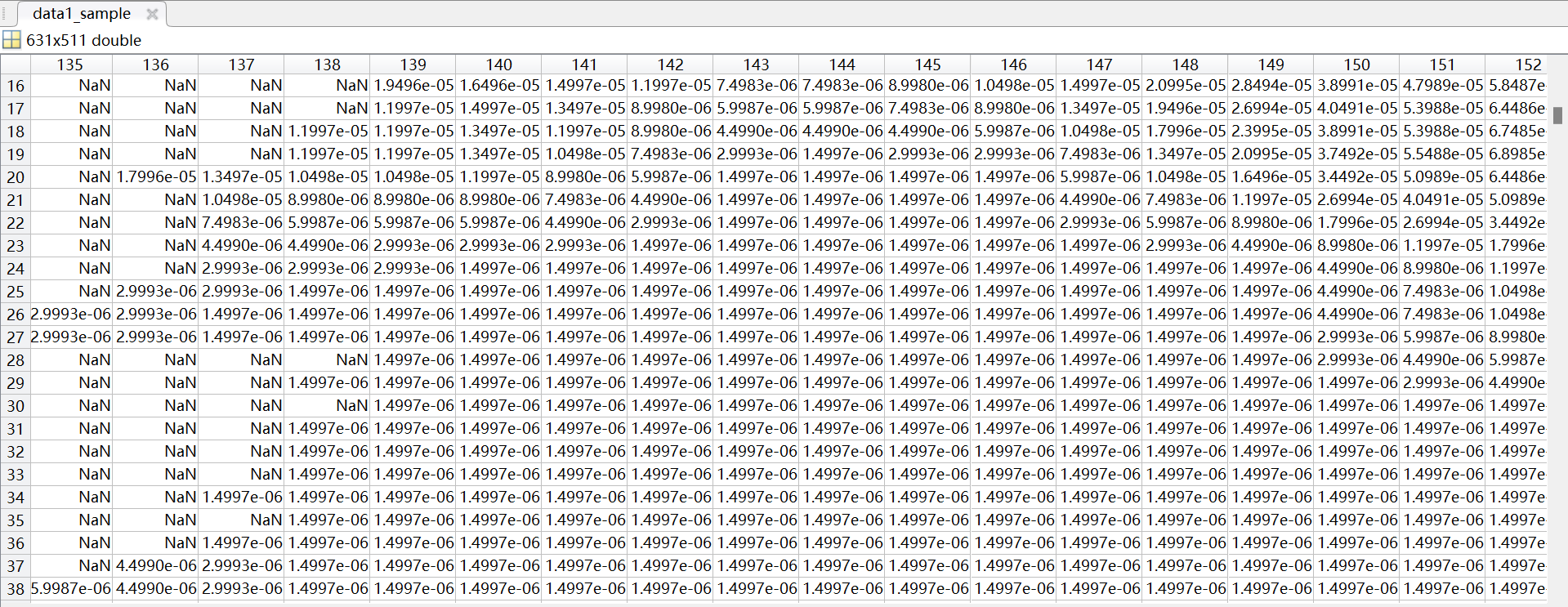
The total precipitation data for all hours in January 1970 that I downloaded is a 197001.nc file, and I want to split it into an NC file for each day of January, but the TP value in the split 19700101.nc is inconsistent with the TP value in the total precipitation hour file 19700101.nc for January 1, 1970 that I downloaded directly, and the TP value in the split NC file has a negative value. How can I properly split 197001.nc file into NC file for each day of the month?
In MATLAB, I read the code for the NC file as follows:
clc;
clear;
filename = ‘E:\a.code\yy\yy_pythoncode\chinanc\test\out\1970.01.01.nc’;% The January 1, 1970 NC file is a January 1, 1970 NC file that was split from the hourly NC file of accumulated precipitation in January 1970 using Python
data = ncread(filename, ‘tp’);
data_sample=data(:,:,1);
data_sample(data_sample ==-1.977711546252703e-06) = NaN;
filename = ‘E:\a.code\yy\yy_pythoncode\chinanc\test\xiari\chinanc\tp19700101.nc’;% The NC file of accumulated precipitation on January 1, 1970, which can be downloaded directly from the official website
data1 = ncread(filename, ‘tp’);
data1_sample=data1(:,:,1);
data1_sample(data1_sample ==-1.977711546252703e-06) = NaN;
Below is a Python code to split the hourly NC file of the accumulated precipitation for January 1970 into an hourly NC file for each day:
import netCDF4 as nc
import numpy as np
import os
Read monthly files
monthly_file = r’E:\a.code\yy\yy_pythoncode\chinanc\test\yuan\1970-01.nc’
dataset = nc.Dataset(monthly_file)
Extract time and tp variables
time_var = dataset.variables[‘time’]
tp_var = dataset.variables[‘tp’]
Determine the index range for each day ( a time step of 1 hour)
time_step_per_day = 24
Output directory
output_dir = r’E:\a.code\yy\yy_pythoncode\chinanc\test\out’
os.makedirs(output_dir, exist_ok=True)
Cycle through each day, extracting and saving daily data
for day in range(31): # there are 31 days in January 1970
start_idx = day * time_step_per_day
end_idx = start_idx + time_step_per_day
daily_tp = tp_var[start_idx:end_idx, :, :]
# Create a new NC file to save the daily data
daily_file = os.path.join(output_dir, f’1970.01.{day+1:02d}.nc’)
with nc.Dataset(daily_file, ‘w’, format=‘NETCDF4’) as daily_dataset:
# Create dimensions and variables
daily_dataset.createDimension(‘time’, time_step_per_day)
daily_dataset.createDimension(‘latitude’, tp_var.shape[1])
daily_dataset.createDimension(‘longitude’, tp_var.shape[2])
time = daily_dataset.createVariable(‘time’, ‘f4’, (‘time’,))
lat = daily_dataset.createVariable(‘latitude’, ‘f4’, (‘latitude’,))
lon = daily_dataset.createVariable(‘longitude’, ‘f4’, (‘longitude’,))
tp = daily_dataset.createVariable(‘tp’, ‘f4’, (‘time’, ‘latitude’, ‘longitude’), fill_value=np.nan)
# Set variable properties that are not _FillValue
for var_name, var in zip([‘time’, ‘latitude’, ‘longitude’, ‘tp’], [time, lat, lon, tp]):
source_var = dataset.variables[var_name if var_name != ‘tp’ else ‘tp’]
for attr_name in source_var.ncattrs():
if attr_name != ‘_FillValue’:
var.setncattr(attr_name, source_var.getncattr(attr_name))
# Copy the data
time[:] = time_var[start_idx:end_idx]
lat[:] = dataset.variables[‘latitude’][:]
lon[:] = dataset.variables[‘longitude’][:]
tp[:] = daily_tp