I’d like to share the revision for the existing official tutorial which will not work with new CDS-beta API and datasets.
Revised tutorial based on Observing floods with GloFAS (link)
Prerequisite
- Installed Python 3.11 or above
- Installed PyCharm
- Installed Miniconda (https://docs.anaconda.com/miniconda/)
- Installed and configured .cdsapi
Setup your environment
Before we begin we need to prepare our environment. This includes:
- Installing the Application Programming Interface (API) of the CDS
- Importing the various python libraries that we will need
Create a new conda environment from a PyCharm project
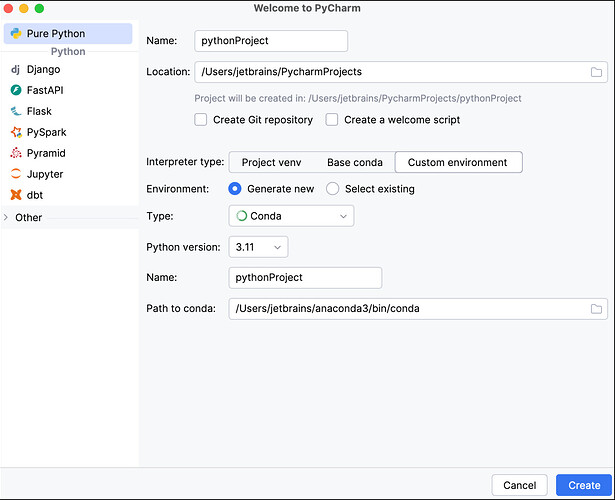
- After opening PyCharm, click New Project.
- In the New Project screen, create the project Name and its Location.
- Select Custom environment, then select Generate New.
- From the Type dropdown, select Conda.
- From the Python version dropdown, select the Python version you want.
- Type the environment name under Name
- Click Create to create the PyCharm project and conda environment.
You can check that the conda environment was created by using conda info --envs in PyCharm’sTerminal
conda info --envs
# conda environments:
base * C:\Users\doc\anaconda3
pythonProject1 C:\Users\doc\anaconda3\envs\pythonProject1
Now you can create a new python file in PyCharm.
Then run following commands in terminal:
Install CDS API
To install the CDS API, run the following command.
pip -q install cdsapi
Install cfgrib and xarray packages
The easiest way to get everything installed is to use conda. To install xarray and cfgrib with its recommended dependencies using the conda command line tool:
conda install -c conda-forge xarray dask netCDF4 bottleneck
conda install -c conda-forge cfgrib
You can install other packages wit pip
pip -q install matplotlib
pip -q install cartopy
Code
Now Insert following code to the newly created python file in PyCharm
# Observing major flood events with GloFAS
# Based on https://ecmwf-projects.github.io/copernicus-training-c3s/glofas-bangladesh-floods.html
#
#
# The tutorial is structured as follows:
#
# 0 Set up your environment (Install and import the required python packages)
# 1 Download the data of interest from the C3S Climate Data Store (CDS);
# 2 Explore the data with xarray, apply mask using the auxiliary data,
# and produce demonstrative map plots;
# 3 Compute and plot time-series of the June 2022 river discharge against
# the 10-year period;
# 4 Compute and display a map of the number of days in June 2022 that the
# river discharge exceeded the 90th percentile of the 10-year period.
#
#
# Creating a new conda environment from a PyCharm project
# 1 After opening PyCharm, click New Project.
# 2 In the New Project screen, create the project Name and its Location.
# 3 Select Custom environment, then select Generate New.
# 4 From the Type dropdown, select Conda.
# 5 From the Python version dropdown, select the Python version you want.
# 6 Type the environment name under Name
# 7 Click Create to create the PyCharm project and conda environment.
# Import libraries
import os
# CDS API
import cdsapi
# Libraries for working with multidimensional arrays
import numpy as np
import xarray as xr
# import cfgrib
# Libraries for plotting and visualising data
import matplotlib.pyplot as plt
import cartopy.crs as ccrs
import cartopy.feature as cfeature
# Disable warnings for data download via API
import urllib3
# Disable xarray runtime warnings
import warnings
urllib3.disable_warnings()
warnings.simplefilter("ignore", category=RuntimeWarning)
# Enter your CDS API key
# If you have not set up your CDS API credentials with a ~/.cdsapirc file,
# it is possible to provide the credentials when initialising the cdsapi.Client.
# replace the ### in the KEY with your real key
if os.path.isfile("~/.cdsapirc"):
cdsapi_kwargs = {}
else:
URL = 'https://cds-beta.climate.copernicus.eu/api'
KEY = '########-####-####-####-###########'
cdsapi_kwargs = {
'url': URL,
'key': KEY,
}
DATADIR = os.path.abspath('./data_dir')
# DATADIR = './data_dir'
os.makedirs(DATADIR, exist_ok=True)
download_file = f"{DATADIR}/glofas-2012_2024.grib"
if not os.path.isfile(download_file):
c = cdsapi.Client(
**cdsapi_kwargs
)
c.retrieve(
'cems-glofas-historical',
{
'system_version': ['version_4_0'],
'hydrological_model': ['lisflood'],
'product_type': ['consolidated'],
'variable': ['river_discharge_in_the_last_24_hours'],
'hyear': [f"{year}" for year in range(2012, 2024)],
'hmonth': ['06'],
'hday': [f"{day:02d}" for day in range(1, 31)],
'format': 'grib2',
'area': [30, 85, 20, 95,],
},
).download(download_file)
glofas_data = xr.open_dataset(download_file, engine='cfgrib')
# print(glofas_data)
# Getting an overview of the study area
# Create a simple plotting function that we can use throughout this notebook
# Define the output plot file path
plot_file = os.path.join(DATADIR, "mean_dis24_june_2022.png")
# print(plot_file)
def plot_map(
plot_data,
title='',
cbar_label='',
cmap='PuBu',
extent=[85, 95, 20, 30],
save_path=None,
**pcolorkwargs
):
# Populate the title and cbar_label with attributes from the plot_data if they have not been
# explicitly specified
title = title or plot_data.attrs.get('long_name', title)
cbar_label = cbar_label or plot_data.attrs.get('units', cbar_label)
# Create a figure with a cartopy projection assigned which allows plotting geospatial data
fig, ax = plt.subplots(
1, 1, figsize=(18, 9), subplot_kw={'projection': ccrs.PlateCarree()}
)
# Plot the data on our figure
im = ax.pcolormesh(
plot_data.longitude, plot_data.latitude, plot_data, cmap=cmap, **pcolorkwargs
)
# Add some additional features
ax.set_title(title, fontsize=16)
ax.gridlines(draw_labels=False, linewidth=1, color='gray', alpha=0.5, linestyle='--')
ax.coastlines(color='black')
# Add country borders in black
ax.add_feature(cfeature.BORDERS, edgecolor='black', lw=1.5, ls=":")
# Set the plot domain/extent
ax.set_extent(extent, crs=ccrs.PlateCarree())
# Add a colour bar
cbar = plt.colorbar(im, fraction=0.04, pad=0.01)
cbar.set_label(cbar_label, fontsize=12)
# Save the graph if a path is specified
if save_path:
plt.savefig(save_path, dpi=300, bbox_inches='tight')
print(f"The graph is saved in {save_path}")
plt.show()
# Calculate the average water consumption for June 2022
mean_data = glofas_data.dis24.mean(dim='time')
mean_data = mean_data.assign_attrs(**glofas_data.dis24.attrs)
# Create and save a graph
plot_map(
mean_data,
vmax=40e3,
vmin=0,
cbar_label="м³ с⁻¹",
save_path=plot_file
)
# Accessing the upstream area data
# choose a target location to download the file:
upstream_area_fname = f"uparea_glofas_v4_0.nc"
upstream_area_file = os.path.join(DATADIR, upstream_area_fname)
# If we have not already downloaded the file, we download it
if not os.path.isfile(upstream_area_file):
u_version = 2 # file version
upstream_data_url = (
f"https://confluence.ecmwf.int/download/attachments/242067380/{upstream_area_fname}?"
f"version={u_version}&modificationDate=1668604690076&api=v2&download=true"
)
import requests
result = requests.get(upstream_data_url)
with open(upstream_area_file, 'wb') as f:
f.write(result.content)
# Open the file and print the contents
upstream_area = xr.open_dataset(upstream_area_file)
# print(upstream_area)
# Reduce the Upstream area data to the domain of the river discharge
# Get the latitude and longitude limits of the data
lat_limits = [glofas_data.latitude.values[i] for i in [0, -1]]
lon_limits = [glofas_data.longitude.values[i] for i in [0, -1]]
up_lats = upstream_area.latitude.values.tolist()
up_lons = upstream_area.longitude.values.tolist()
lat_slice_index = [
round((i-up_lats[0])/(up_lats[1]-up_lats[0]))
for i in lat_limits
]
lon_slice_index = [
round((i-up_lons[0])/(up_lons[1]-up_lons[0]))
for i in lon_limits
]
# Slice upstream area to bangladesh region:
red_upstream_area = upstream_area.isel(
latitude=slice(lat_slice_index[0], lat_slice_index[1]+1),
longitude=slice(lon_slice_index[0], lon_slice_index[1]+1),
)
# There are very minor rounding differences, so we update with the lat/lons from the glofas data
red_upstream_area = red_upstream_area.assign_coords({
'latitude': glofas_data.latitude,
'longitude': glofas_data.longitude,
})
# Add the upstream area to the main data object and print the updated glofas data object:
glofas_data['uparea'] = red_upstream_area['uparea']
# print(glofas_data)
# Mask the river discharge data
glofas_data_masked = glofas_data.where(glofas_data.uparea >= 250.e6)
mean_masked_data = glofas_data_masked.dis24.mean(dim='time').assign_attrs(glofas_data_masked.dis24.attrs)
plot_mask_file = os.path.join(DATADIR, "mean_mean_masked_data.png")
plot_map(
mean_masked_data,
vmax=40e3, vmin=0,
cbar_label="m³ s⁻¹",
save_path=plot_mask_file
)
# Compare the June 2022 river discharge to the previous 10 years
# select the 2012-2021 data
glofas_historical_period = glofas_data_masked.sel(time=slice("2012", "2021"))
glofas_dis24_domain_mean = glofas_historical_period.dis24.mean(["longitude", "latitude"])
# calculate the mean discharge over the whole domain, for each day in June
glofas_xy_climatology = glofas_dis24_domain_mean.groupby('time.day').mean("time")
# calculate the minimum and maximum discharge over the whole domain, for each day in June
glofas_xy_clim_min = glofas_dis24_domain_mean.groupby('time.day').min("time")
glofas_xy_clim_max = glofas_dis24_domain_mean.groupby('time.day').max("time")
# Calculate the 10th and 90th percentiles of discharge rate of the whole domain
glofas_xy_clim_quantiles = glofas_dis24_domain_mean.groupby('time.day').quantile(dim="time", q=[0.1, 0.9])
# calculate the mean discharge over the whole domain for each day in June 2022
glofas_xy_2022 = glofas_data_masked.dis24.sel(time="2022")
glofas_xy_2022_mean = glofas_xy_2022.mean(['longitude', 'latitude']).groupby('time.day').mean("time")
# Create a figure and axis
fig = plt.figure(figsize=(8, 6))
ax = fig.add_subplot()
# Plot the minimum to maximum range
ax.fill_between(
np.arange(1, 31), glofas_xy_clim_min.values, glofas_xy_clim_max.values,
color="k", alpha=0.2, label='min/max percentile range'
)
# Plot the 10th to 90th percentile range
ax.fill_between(
np.arange(1, 31),
glofas_xy_clim_quantiles.isel(quantile=0).values,
glofas_xy_clim_quantiles.isel(quantile=1).values,
color="k", alpha=0.5, label='10th/90th percentile range'
)
# Plot the mean for the 10 year period
(line_base,) = plt.plot(
np.arange(1, 31), glofas_xy_climatology.values, color="k", label="Reference period mean"
)
# Plot the region where 2022 is greater than the mean in blue
ax.fill_between(
range(1, int(glofas_xy_2022_mean.shape[0]) + 1),
glofas_xy_climatology.values[: int(glofas_xy_2022_mean.shape[0])],
glofas_xy_2022_mean.values,
color="b",
alpha=0.4,
where=glofas_xy_2022_mean.values >= glofas_xy_climatology.values[: int(glofas_xy_2022_mean.shape[0])],
interpolate=True, label="2022 excess discharge"
)
# Plot the region where 2022 is less than the mean in red
ax.fill_between(
range(1, int(glofas_xy_2022_mean.shape[0]) + 1),
glofas_xy_climatology.values[: int(glofas_xy_2022_mean.shape[0])],
glofas_xy_2022_mean.values,
color="r",
alpha=0.4,
where=glofas_xy_2022_mean.values < glofas_xy_climatology.values[: int(glofas_xy_2022_mean.shape[0])],
interpolate=True,
)
# Plot the 2022 data points
dots = ax.scatter(
range(1, int(glofas_xy_2022_mean.shape[0]) + 1),
glofas_xy_2022_mean.values,
color="k",
# label="2022",
zorder=2,
)
# Add a legend and axis labels
ax.legend(loc='upper left')
plt.xlabel("Day in June")
plt.ylabel("Daily river discharge ($m^{3}s^{-1}$)")
# Save the plot
plot_save_path = os.path.join(DATADIR, "june_2022_discharge_comparison.png")
plt.savefig(plot_save_path, dpi=300, bbox_inches='tight')
plt.close() # Close the plot to free up memory
# Calculate the climatology for the historical period: 2012-2021
glofas_historical_period = glofas_data_masked.sel(time=slice("2012", "2021"))
# glofas_climatology = glofas_data_masked.groupby('time.dayofyear').mean("time")
glofas_climat_quantiles = glofas_historical_period.groupby('time.dayofyear').quantile(dim="time", q=[0.1, 0.9])
# # Calculate the anomaly w.r.t the above climatology
# glofas_anomaly = glofas_data_masked.groupby('time.dayofyear') - glofas_climatology
# # Calculate this a relative anomaly
# glofas_relanom = glofas_anomaly.groupby('time.dayofyear')/glofas_climatology
glofas_high = glofas_data_masked.groupby('time.dayofyear') > glofas_climat_quantiles.sel(quantile=0.9)
glofas_high = glofas_high.where(glofas_data.uparea > 250e6)
# Plot the data:
year = '2022'
number_of_high_days = glofas_high.sel(time=year).dis24.sum(dim='time')
number_of_high_days = number_of_high_days.assign_attrs(glofas_high.dis24.attrs)
plot_map_file = os.path.join(DATADIR, "map_river_discharge.png")
plot_map(
number_of_high_days,
vmax=25,
cmap='Purples',
cbar_label='Number of days',
save_path=plot_map_file
)