Dear all,
I downloaded CAMS data from global atmospheric composition forecast for a simulation with WRF-Chem following the instructions How to run the WRF-Chem model using CAMS data as initial and boundary conditions (BC)?#Step4Runningthemodel
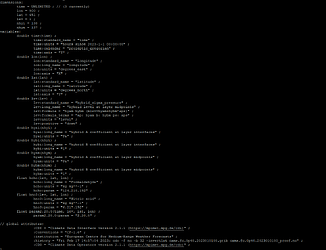
The process of downloading the grib data with a python request, converting it to netcdf using the script of section 2.2 of the instructions worked well for 2021-08-01 data. So, I could follow next instructions to finally run mozbc and wrf.exe incorporating successfully BC from CAMS to WRF-Chem.
However, when I downloaded same kind of data for 2022 or 2023 (python request attached before) I got problems converting grib data to netcdf. Specifically running the command cdo -f nc -b 32 -invertlat GRG_XXXX.grib GRG_XXXX.netcdf
- I got this warning and netcdf output.

- There is a problem with parameter pan, which is not defined at timestep 1, although I retrieved and downloaded this parameter inside the same request as the other 7 parameters (see the python request attached)
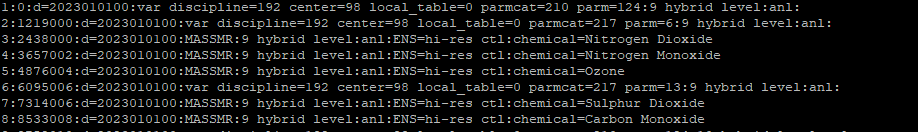
- It seems that only 2 of 8 variables were correctly recognized but time dimension were not incorporated for these two variables.
- Last parameter converted only show param2.20.0. Then I noticed that the grib format of certain chemical parameters changed from mid 2021 between grib edition 1 to grib edition 2https://apps.ecmwf.int/codes/grib/param-db?id=210121
- Then I downloaded the last version of cdo but it didn't also recognize the parameter NO2. I also attempted to create my own Parameter table (gribtab) as follows according to the wiki: https://code.zmaw.de/projects/cdo/embedded/index.html#x1-300001.6 but I got probelms with the id for NO and NO2 which are the same in grib2 edition.
- Finally I tried to convert grib to netcdf using last eccode version with command grib_to_netcdf. This command worked well recognising the parameters but it didn't incorporate the hybrid coordinates hyai, hybi, hyam, hybm to the netcdf which are essential to follow next steps and run mozbc and wrf.exe.
I wonder how could I convert grib data to netcdf for NO2, NO, CO, SO2, pan, CH2O CAMS forecast composition chemical species (for 2022 and 2023), obtaining all the parameters with 4 dimensions (time, lev, lat, lon) and the hybrid coordinates hyai, hybi, hyam, hybm in the same output nc file.
Many thanks,
Kind regards,
Maria